See also More Shock and Awe: James Lee and SSGAC in Oslo, 600 SNP hits.
Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits and implications for the future
Yan Zhang, Guanghao Qi, Ju-Hyun Park, Nilanjan Chatterjee (Johns Hopkins University)
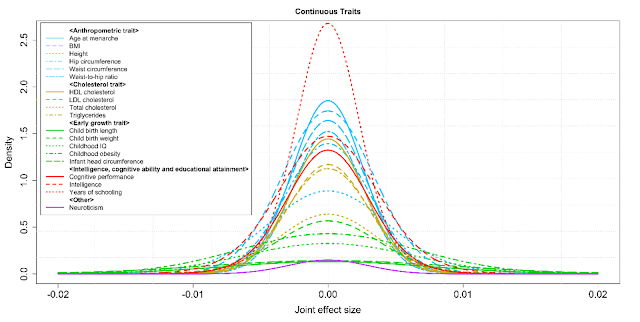
Summary-level statistics from genome-wide association studies are now widely used to estimate heritability and co-heritability of traits using the popular linkage-disequilibrium-score (LD-score) regression method. We develop a likelihood-based approach for analyzing summary-level statistics and external LD information to estimate common variants effect-size distributions, characterized by proportion of underlying susceptibility SNPs and a flexible normal-mixture model for their effects. Analysis of summary-level results across 32 GWAS reveals that while all traits are highly polygenic, there is wide diversity in the degrees of polygenicity. The effect-size distributions for susceptibility SNPs could be adequately modeled by a single normal distribution for traits related to mental health and ability and by a mixture of two normal distributions for all other traits. Among quantitative traits, we predict the sample sizes needed to identify SNPs which explain 80% of GWAS heritability to be between 300K-500K for some of the early growth traits, between 1-2 million for some anthropometric and cholesterol traits and multiple millions for body mass index and some others. The corresponding predictions for disease traits are between 200K-400K for inflammatory bowel diseases, close to one million for a variety of adult onset chronic diseases and between 1-2 million for psychiatric diseases.
This figure shows predicted effect size distributions for a number of quantitative traits. You can see that height and intelligence are somewhat different, but not dramatically so.


No comments:
Post a Comment